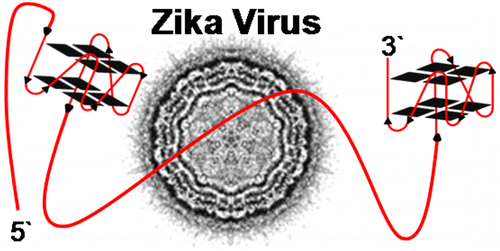
The rapid spread of the Zika virus has researchers working to find vulnerabilities in the virus and develop a treatment. Chemists Cynthia Burrows and Aaron Fleming used DNA structures called G-quadruplexes as genetic landmarks to compare Zika with other related viruses. In a new study, Burrows and Fleming describe how the pattern of G-quadruplexes in Zika’s genome suggests a possible drug target.
Abstract

Zika virus has emerged as a global concern because neither a vaccine nor antiviral compounds targeting it exist. A structure for the positive-sense RNA genome has not been established, leading us to look for potential G-quadruplex sequences (PQS) in the genome. The analysis identified >60 PQSs in the Zika genome. To minimize the PQS population, conserved sequences in the Flaviviridae family were found by sequence alignment, identifying seven PQSs in the prM, E, NS1, NS3, and NS5 genes. Next, alignment of 78 Zika strain genomes identified a unique PQS near the end of the 3′-UTR. Structural studies on the G-quadruplex sequences found four of the conserved Zika virus sequences to adopt stable, parallel-stranded folds that bind a G-quadruplex-specific compound, and one that was studied caused polymerase stalling when folded to a G-quadruplex. Targeting these PQSs with G-quadruplex binding molecules validated in previous clinical trials may represent a new approach for inhibiting viral replication.
Read the article from ACS Infectious Diseases here: http://pubs.acs.org/doi/ipdf/10.1021/acsinfecdis.6b00109
